Multi-Run (i.e. sets) Example of mzQC
Here, we describe an mzQC JSON document used to convey QC data which is computed on a set of runs, i.e.
is only interpretable in the context of this set (group).
Of course, QC metrics which refer to each run individually can also be stored, also in the same mzQC file
(see our example individual-runs.mzQC.md on how to do that), but this example is about group/set metrics.
Find the complete example file at the bottom of this document or in the example folder.
The basic structure of our mzQC file is identical to the individual-runs.mzQC example, i.e.
the documents main anchor is between the outer curly brackets:
{ "mzQC":
{
...
}
}
Within this main anchor, there are usually the following sections: a) general information about the file,
"version": "1.0.0",
"creationDate": "2020-12-21T11:56:34",
"contactName": "Chris Bielow",
"contactAddress": "chris.bielow@bsc.fu-berlin.de",
"description": "A simple mzQC file containing information for sets of runs.",
b) reference information for controlled vocabularies (cv) at the bottom,
"controlledVocabularies": [
{
"name": "Proteomics Standards Initiative Quality Control Ontology",
"uri": "https://github.com/HUPO-PSI/qcML-development/blob/master/cv/v0_1_0/qc-cv.obo",
"version": "0.1.0"
},
{
"name": "Proteomics Standards Initiative Mass Spectrometry Ontology",
"uri": "https://github.com/HUPO-PSI/psi-ms-CV/blob/master/psi-ms.obo",
"version": "4.1.7"
}
]
and (now in addition or as replacement) to the runQualities of the individual-runs.mzQC we have
c) information about the QC metrics computed on a set of runs.
"setQualities": [
{
...
}
]
In fact, setQualities can contain one or more setQuality objects, each defining a different set of runs.
E.g. if you have three technical replicates for two conditions for at total of six runs, you might want to subsume three runs into a set, one for each condition and report the total number of proteins you identified, or the percentage of total intensity attributable to contaminants). Each setQuality object is an element of a JSON array, thus it is not explicitly named (i.e. there is no “setQuality” key in the mzQC file).
For the purpose of this example, we will use three setQuality objects (there could be none, only one or more than two though):
the **healthy** set: tr1_healthy, tr2_healthy, tr3_healthy
the **diseased** set: tr1_diseased, tr2_diseased, tr3_diseased
the **all** set: tr1_healthy, tr2_healthy, tr3_healthy, tr1_diseased, tr2_diseased, tr3_diseased
How you define (and name) each set, is up to you and depends on your experimental design and the kind of comparisons you want to make.
A setQuality represents QC data that must be viewed in the context of all the runs of this set/group. I.e. the data is only valid within the context of the runs it comprises. E.g. it would be invalid to define a set of three runs and report their individual MS1 scan counts as a 3-tuple – because this information can clearly be attributed to individual runs and thus belongs in three separate runQuality objects, rather than a single setQuality.
Similar to runQuality, a setQuality also contains metadata about the set of runs (its input files, the software used, etc).
You can give the set a unique name using the label attribute. Here is how a setQuality object looks like:
{
"metadata": {
"label": "healthy"
"inputFiles":
...
},
"qualityMetrics": [
...
]
}
The inputFiles consist of an array of inputFile objects, describing the source files with structured information about the file’s name, format, location and other properties, defined via cv terms.
"inputFiles": [
{
"name": "tr1_healthy",
"location": "c:\msdata\techRep1_healthy.mzML",
...
},
{
"name": "tr2_healthy",
"location": "c:\msdata\techRep2_healthy.mzML",
...
},
{
"name": "tr3_healthy",
"location": "c:\msdata\techRep3_healthy.mzML",
...
}
]
The inputFile object is only sketched here. It can contain a lot more information, such as file format and further properties. See the full example below or individual-runs.mzQC for details.
In qualityMetrics, we will store the actual QC information for a particular setQuality. Each qualityMetric has an accession and the corresponding name as defined by the QC controlled vocabulary (see qc-cv.obo). They should be represented exactly as stated in the .obo file. The value carries the actual information and can be either a single value, a tuple of values, a matrix or table. Below, we will look at single values and tables.
Lets start with our first metric Protein contaminant intensity ratio. It describes the relative intensity (in [0, 1]) of all contaminant proteins (from all runs in the set) – the higher the value the more contaminants are present in the runs of the set.
"accession": "QC:0000000",
"name": "Protein contaminant intensity ratio",
"value": 0.25
We compute this metric for each set, in our case for the healthy as well as the diseased set, but not for the all set (because we want to keep the example small). But in general, what metrics you compute is up to you.
Our second example is a principal component analysis (PCA) result matrix.
The setQuality where this PCA metric will be stored, references all runs as input files.
The input table for a PCA computation can be found, for example, in MaxQuant’s proteinGroups.txt output file. To stick with this example, the table in proteinGroups.txt has rows (proteins) and columns (groups, e.g. healthy or diseased), and the values in the table are protein abundances. Thus, MaxQuant has already aggregated the data from rawfiles(=runs) belonging to a certain group for us (e.g. by averaging the protein abundances). Now your QC software can derive a new table using PCA, where each group is represented by PCA coordinates.
First, let’s see what the PCA plot would look like:
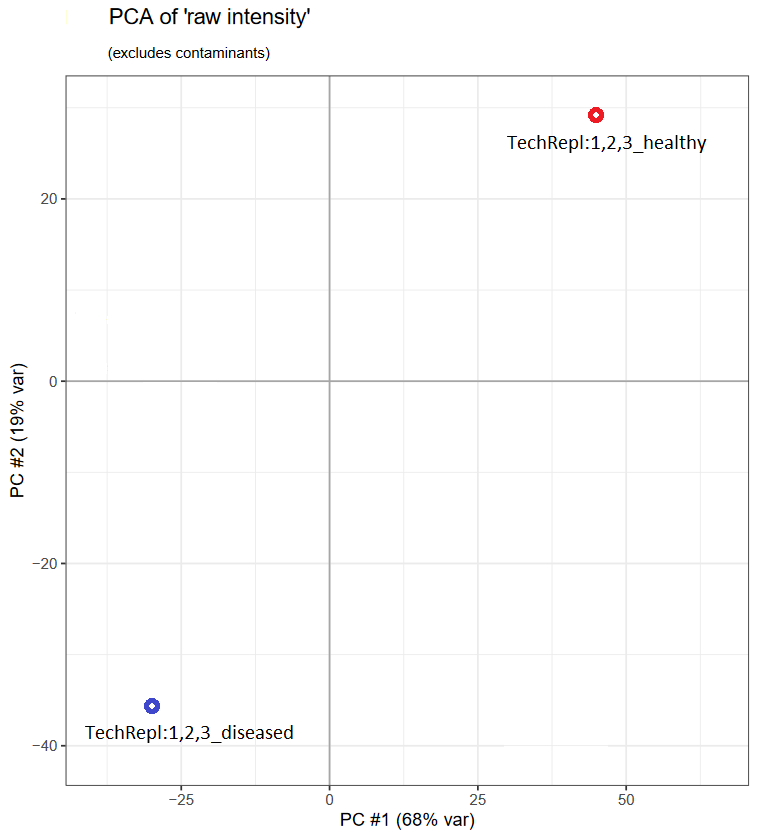 Now, let’s look at the mzQC data which allows to create this plot: We use two separate metrics. One named
Now, let’s look at the mzQC data which allows to create this plot: We use two separate metrics. One named group of runs to associate runs to groups, and secondly a PCA table metric to store the PCA data (the first 5 principal components for each group).
"setQualities": [
...,
{
...,
"qualityMetrics": [
{
"accession": "QC:4000264",
"name": "group of runs",
"value": {
"inputfile_name": ["tr1_healthy", "tr2_healthy", "tr3_healthy" , "tr1_diseased", "tr2_diseased", "tr3_diseased"],
"group-label": ["healthy" , "healthy" , "healthy" , "diseased" , "diseased" , "diseased"]
}
},
{
"accession": "QC:4000267",
"name": "PCA table",
"value": {
"group-label": ["healthy", "diseased"],
"PCA Dimension 1": [47.22, -30.22],
"PCA Dimension 2": [29.1, -36.5],
"PCA Dimension 3": [3.8, -7.3],
"PCA Dimension 4": [-7.7, 5.55],
"PCA Dimension 5": [140.6, -64.1]
}
}
}
]
]
Note: the group of runs metric can be defined only once per setQuality, but can be referenced in many metrics (here, for our PCA table) in that context.
If you look closely, we somewhat defined the group healthy twice. Once as an individual setQuality and once via the group of runs qualityMetric in the all set.
There is no easy way around this. If we were to omit the all set, we’d need to distribute the columns of the PCA table metric into separate setQuality objects (and whoever wants to plot it, needs to puzzle it back together; not ideal).
On the other hand, ommitting the healthy/diseased setQualities is not sensible either, because then there would be only the all setQuality where all data for different subsets would need to reside.
This is the mzQC file once again, in full:
{
"mzQC": {
"version": "1.0.0",
"creationDate": "2020-12-01T14:19:09",
"contactName": "Chris Bielow",
"contactAddress": "chris.bielow@bsc.fu-berlin.de",
"description": "A simple mzQC file containing information for sets of runs.",
"setQualities": [
{
"metadata": {
"label": "healthy",
"inputFiles": [
{
"name": "tr1_healthy",
"location": "c:\\msdata\\techRep1_healthy.mzML",
"fileFormat": {
"accession": "MS:1000584",
"name": "mzML format"
},
"fileProperties": [
{
"accession": "MS:1000747",
"name": "completion time",
"value": "2012-02-03 11:00:41"
}
]
},
{
"name": "tr2_healthy",
"location": "c:\\msdata\\techRep2_healthy.mzML",
"fileFormat": {
"accession": "MS:1000584",
"name": "mzML format"
},
"fileProperties": [
{
"accession": "MS:1000747",
"name": "completion time",
"value": "2012-02-03 13:00:41"
}
]
},
{
"name": "tr3_healthy",
"location": "c:\\msdata\\techRep3_healthy.mzML",
"fileFormat": {
"accession": "MS:1000584",
"name": "mzML format"
},
"fileProperties": [
{
"accession": "MS:1000747",
"name": "completion time",
"value": "2012-02-03 14:00:41"
}
]
}
],
"analysisSoftware": [
{
"accession": "MS:1001058",
"name": "quality estimation by manual validation",
"version": "0",
"uri": "https://dx.doi.org/10.1021/pr201071t"
}
]
},
"qualityMetrics": [
{
"accession": "QC:0000000",
"name": "Protein contaminant intensity ratio",
"value": "0.25"
}
]
},
{
"metadata": {
"label": "diseased",
"inputFiles": [
{
"name": "tr1_diseased",
"location": "c:\\msdata\\techRep1_diseased.mzML",
"fileFormat": {
"accession": "MS:1000584",
"name": "mzML format"
},
"fileProperties": [
{
"accession": "MS:1000747",
"name": "completion time",
"value": "2012-02-03 12:00:41"
}
]
},
{
"name": "tr2_diseased",
"location": "c:\\msdata\\techRep2_diseased.mzML",
"fileFormat": {
"accession": "MS:1000584",
"name": "mzML format"
},
"fileProperties": [
{
"accession": "MS:1000747",
"name": "completion time",
"value": "2012-02-03 14:00:41"
}
]
},
{
"name": "tr3_diseased",
"location": "c:\\msdata\\techRep3_diseased.mzML",
"fileFormat": {
"accession": "MS:1000584",
"name": "mzML format"
},
"fileProperties": [
{
"accession": "MS:1000747",
"name": "completion time",
"value": "2012-02-03 15:00:41"
}
]
}
],
"analysisSoftware": [
{
"accession": "MS:1001058",
"name": "quality estimation by manual validation",
"version": "0",
"uri": "https://dx.doi.org/10.1021/pr201071t"
}
]
},
"qualityMetrics": [
{
"accession": "QC:0000000",
"name": "Protein contaminant intensity ratio",
"value": "0.31"
}
]
},
{
"metadata": {
"label": "all",
"inputFiles": [
{
"name": "tr1_healthy",
"location": "c:\\msdata\\techRep1_healthy.mzML",
"fileFormat": {
"accession": "MS:1000584",
"name": "mzML format"
},
"fileProperties": [
{
"accession": "MS:1000747",
"name": "completion time",
"value": "2012-02-03 11:00:41"
}
]
},
{
"name": "tr2_healthy",
"location": "c:\\msdata\\techRep2_healthy.mzML",
"fileFormat": {
"accession": "MS:1000584",
"name": "mzML format"
},
"fileProperties": [
{
"accession": "MS:1000747",
"name": "completion time",
"value": "2012-02-03 13:00:41"
}
]
},
{
"name": "tr3_healthy",
"location": "c:\\msdata\\techRep3_healthy.mzML",
"fileFormat": {
"accession": "MS:1000584",
"name": "mzML format"
},
"fileProperties": [
{
"accession": "MS:1000747",
"name": "completion time",
"value": "2012-02-03 14:00:41"
}
]
},
{
"name": "tr1_diseased",
"location": "c:\\msdata\\techRep1_diseased.mzML",
"fileFormat": {
"accession": "MS:1000584",
"name": "mzML format"
},
"fileProperties": [
{
"accession": "MS:1000747",
"name": "completion time",
"value": "2012-02-03 12:00:41"
}
]
},
{
"name": "tr2_diseased",
"location": "c:\\msdata\\techRep2_diseased.mzML",
"fileFormat": {
"accession": "MS:1000584",
"name": "mzML format"
},
"fileProperties": [
{
"accession": "MS:1000747",
"name": "completion time",
"value": "2012-02-03 14:00:41"
}
]
},
{
"name": "tr3_diseased",
"location": "c:\\msdata\\techRep3_diseased.mzML",
"fileFormat": {
"accession": "MS:1000584",
"name": "mzML format"
},
"fileProperties": [
{
"accession": "MS:1000747",
"name": "completion time",
"value": "2012-02-03 15:00:41"
}
]
}
],
"analysisSoftware": [
{
"accession": "MS:1001058",
"name": "quality estimation by manual validation",
"version": "0",
"uri": "https://dx.doi.org/10.1021/pr201071t"
}
]
},
"qualityMetrics": [
{
"accession": "QC:4000264",
"name": "group of runs",
"value": {
"inputfile_name": ["tr1_healthy", "tr2_healthy", "tr3_healthy" , "tr1_diseased", "tr2_diseased", "tr3_diseased"],
"group-label": ["healthy" , "healthy" , "healthy" , "diseased" , "diseased" , "diseased"]
}
},
{
"accession": "QC:4000267",
"name": "PCA table",
"value": {
"group-label": ["healthy", "diseased"],
"PCA Dimension 1": [47.22, -30.22],
"PCA Dimension 2": [29.1, -36.5],
"PCA Dimension 3": [3.8, -7.3],
"PCA Dimension 4": [-7.7, 5.55],
"PCA Dimension 5": [140.6, -64.1]
}
}
]
}
],
"controlledVocabularies": [
{
"name": "Proteomics Standards Initiative Quality Control Ontology",
"uri": "https://github.com/HUPO-PSI/qcML-development/blob/master/cv/v0_1_0/qc-cv.obo",
"version": "0.1.0"
},
{
"name": "Proteomics Standards Initiative Mass Spectrometry Ontology",
"uri": "https://github.com/HUPO-PSI/psi-ms-CV/blob/master/psi-ms.obo",
"version": "4.1.7"
}
]
}
}